hit
This module implements class relative to hit and some functions to do some computation on hit objects.
Modelize a hmm hit on the replicon. There is only one Corehit for a CoreGene. |
|
Modelize a hit and its relation to the Model. |
|
Parent class of Loner, MultiSystem. It’s inherits from ModelHit. |
|
Modelize “true” Loner. |
|
Modelize hit which can be used in several Systems (same model) |
|
Modelize a hit representing a gene Loner and MultiSystem at same time. |
|
The weights apply to the hit to compute score |
|
Return the best hit for a given function |
|
Sort hits |
|
Choose among svereal multisystem hits the best one |
|
If several profile hit the same gene return the best hit |
A Hit is created when hmmsearch find similarities between a profile and protein of the input dataset
Below the ingheritance diagram of Hits
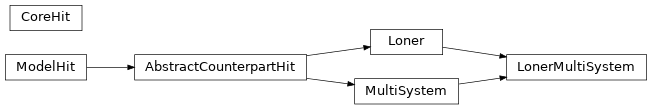
And a diagram showing the interaction between CoreGene, ModelGene, Model, Hit, Loner, … interactions
The diagram above represents the models, genes and hit generated from the definitions below.
<model name="A" inter_gene_max_space="2">
<gene name="abc" presence="mandatory"/>
<gene name="def" presence="accessory"/>
</model>
<model name="B" inter_gene_max_space="5">
<gene name="def" presence="mandatory"/>
<exchangeables>
<gene name="abc"/>
</exchangeables>
<gene name="ghj" presence="accessory"
</model>
hit API reference
CoreHit
- class macsypy.hit.CoreHit(gene, hit_id, hit_seq_length, replicon_name, position_hit, i_eval, score, profile_coverage, sequence_coverage, begin_match, end_match)[source]
Handle the hits filtered from the Hmmer search. The hits are instanciated by
HMMReport.extract()method In one run of MacSyFinder, there exists only one CoreHit per gene These hits are independent of anymacsypy.model.Modelinstance.- __eq__(other)[source]
Return True if two hits are totally equivalent, False otherwise.
- Parameters:
other (
macsypy.report.CoreHitobject) – the hit to compare to the current object- Returns:
the result of the comparison
- Return type:
boolean
- __gt__(other)[source]
compare two Hits. If the sequence identifier is the same, do the comparison on the score. Otherwise, do it on alphabetical comparison of the sequence identifier.
- Parameters:
other (
macsypy.report.CoreHitobject) – the hit to compare to the current object- Returns:
True if self is > other, False otherwise
- __init__(gene, hit_id, hit_seq_length, replicon_name, position_hit, i_eval, score, profile_coverage, sequence_coverage, begin_match, end_match)[source]
- Parameters:
gene (
macsypy.gene.CoreGeneobject) – the gene corresponding to this profilehit_id (str) – the identifier of the hit
hit_seq_length (int) – the length of the hit sequence
replicon_name (str) – the name of the replicon
position_hit (int) – the rank of the sequence matched in the input dataset file
i_eval (float) – the best-domain evalue (i-evalue, “independent evalue”)
score (float) – the score of the hit
profile_coverage (float) – percentage of the profile that matches the hit sequence
sequence_coverage (float) – percentage of the hit sequence that matches the profile
begin_match (int) – where the hit with the profile starts in the sequence
end_match (int) – where the hit with the profile ends in the sequence
- __lt__(other)[source]
Compare two Hits. If the sequence identifier is the same, do the comparison on the score. Otherwise, do it on alphabetical comparison of the sequence identifier.
- Parameters:
other (
macsypy.report.CoreHitobject) – the hit to compare to the current object- Returns:
True if self is < other, False otherwise
- __str__()[source]
- Returns:
Useful information on the CoreHit: regarding Hmmer statistics, and sequence information
- Return type:
str
- __weakref__
list of weak references to the object (if defined)
ModelHit
- class macsypy.hit.ModelHit(hit, gene_ref, gene_status)[source]
Encapsulates a
macsypy.report.CoreHitThis class stores a CoreHit that has been attributed to a putative system. Thus, it also stores:the system,
the status of the gene in this system, (‘mandatory’, ‘accessory’, …
the gene in the model for which it’s an occurrence
for one gene it can exist several ModelHit instance one for each Model containing this gene
- __init__(hit, gene_ref, gene_status)[source]
- Parameters:
hit (
macsypy.hit.CoreHitobject) – a match between a hmm profile and a replicongene_ref (
macsypy.gene.ModelGeneobject) –The ModelGene link to this hit The ModeleGene have the same name than the CoreGene But one hit can be link to several ModelGene (several Model) To know for what gene this hit play role use the
macsypy.gene.ModelGene.alternate_of()hit.gene_ref.alternate_of()
gene_status (
macsypy.gene.GeneStatusobject) –
- __weakref__
list of weak references to the object (if defined)
- property hit
- Returns:
The CoreHit below this ModelHit
- Return type:
macsypy.hit.CoreHitoject
- property loner
- Returns:
True if the hit represent a loner
macsypy.Gene.ModelGene, False otherwise. A True Loner is a hit representing a gene with the attribute loner and which does not include in a cluster.a hit representing a loner gene but include in a cluster is not a true loner
a hit which is not include with other gene in a cluster but does not represent a gene loner is not a True loner (This situation may append when min_genes_required = 1)
- Return type:
bool
- property multi_model
- Returns:
True if the hit represent a multi_model
macsypy.Gene.ModelGene, False otherwise.- Return type:
bool
- property multi_system
- Returns:
True if the hit represent a multi_system
macsypy.Gene.ModelGene, False otherwise.- Return type:
bool
AbstractCounterpartHit
- class macsypy.hit.AbstractCounterpartHit(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
Abstract Class to handle ModelHit wit equivalent for instance Loner or MultiSystem hit
- __init__(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
- Parameters:
hit (
macsypy.hit.CoreHitobject) – a match between a hmm profile and a replicongene_ref (
macsypy.gene.ModelGeneobject) –The ModelGene link to this hit The ModeleGene have the same name than the CoreGene But one hit can be link to several ModelGene (several Model) To know for what gene this hit play role use the
macsypy.gene.ModelGene.alternate_of()hit.gene_ref.alternate_of()
gene_status (
macsypy.gene.GeneStatusobject) –
- property counterpart
- Returns:
The set of hits that can play the same role
- property loner
- Returns:
True if the hit represent a loner
macsypy.Gene.ModelGene, False otherwise. A True Loner is a hit representing a gene with the attribute loner and which does not include in a cluster.a hit representing a loner gene but include in a cluster is not a true loner
a hit which is not include with other gene in a cluster but does not represent a gene loner is not a True loner (This situation may append when min_genes_required = 1)
- Return type:
bool
- property multi_system
- Returns:
True if the hit represent a multi_system
macsypy.Gene.ModelGene, False otherwise.- Return type:
bool
Loner
- class macsypy.hit.Loner(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
Handle hit which encode for a gene tagged as loner and which not clustering with other hit.
- __init__(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
hit that is outside a cluster, the gene_ref is a loner
- Parameters:
hit (
macsypy.hit.CoreHitobject) – a match between a hmm profile and a replicongene_ref (
macsypy.gene.ModelGeneobject) –The ModelGene link to this hit The ModeleGene have the same name than the CoreGene But one hit can be link to several ModelGene (several Model) To know for what gene this hit play role use the
macsypy.gene.ModelGene.alternate_of()hit.gene_ref.alternate_of()
gene_status (
macsypy.gene.GeneStatusobject) –counterpart (list of
macsypy.hit.CoreHit) – the other occurence of the gene or exchangeable in the replicon
- property loner
- Returns:
True if the hit represent a loner
macsypy.Gene.ModelGene, False otherwise. A True Loner is a hit representing a gene with the attribute loner and which does not include in a cluster.a hit representing a loner gene but include in a cluster is not a true loner
a hit which is not include with other gene in a cluster but does not represent a gene loner is not a True loner (This situation may append when min_genes_required = 1)
- Return type:
bool
MultiSystem
- class macsypy.hit.MultiSystem(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
Handle hit which encode for a gene tagged as loner and which not clustering with other hit.
- __init__(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
hit that is outside a cluster, the gene_ref is a loner
- Parameters:
hit (
macsypy.hit.CoreHitobject) – a match between a hmm profile and a replicongene_ref (
macsypy.gene.ModelGeneobject) –The ModelGene link to this hit The ModeleGene have the same name than the CoreGene But one hit can be link to several ModelGene (several Model) To know for what gene this hit play role use the
macsypy.gene.ModelGene.alternate_of()hit.gene_ref.alternate_of()
gene_status (
macsypy.gene.GeneStatusobject) –counterpart (list of
macsypy.hit.CoreHit) – the other occurence of the gene or exchangeable in the replicon
- property multi_system
- Returns:
True if the hit represent a multi_system
macsypy.Gene.ModelGene, False otherwise.- Return type:
bool
LonerMultiSystem
- class macsypy.hit.LonerMultiSystem(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
- Handle hit which encode for a gene
gene tagged as multi-system
and gene tagged as loner also
and the hit do not clustering with other hits.
- __init__(hit, gene_ref=None, gene_status=None, counterpart=None)[source]
hit that is outside a cluster, the gene_ref is loner and multi_system
- Parameters:
hit (
macsypy.hit.CoreHit|macsypy.hit.ModelHit|macsypy.hit.MultiSystemobject) – a match between a hmm profile and a replicongene_ref (
macsypy.gene.ModelGeneobject) –The ModelGene link to this hit The ModeleGene have the same name than the CoreGene But one hit can be link to several ModelGene (several Model) To know for what gene this hit play role use the
macsypy.gene.ModelGene.alternate_of()hit.gene_ref.alternate_of()
gene_status (
macsypy.gene.GeneStatusobject) –counterpart (list of
macsypy.hit.CoreHit) – the other occurence of the gene or exchangeable in the replicon
HitWeight
- class macsypy.hit.HitWeight(itself: float = 1, exchangeable: float = 0.8, mandatory: float = 1, accessory: float = 0.5, neutral: float = 0, out_of_cluster: float = 0.7)[source]
The weight to compute the cluster and system score see user documentation macsyfinder functionning for further details by default
itself = 1
exchangeable = 0.8
mandatory = 1
accessory = 0.5
neutral = 0
out_of_cluster = 0.7
- __delattr__(name)
Implement delattr(self, name).
- __eq__(other)
Return self==value.
- __hash__()
Return hash(self).
- __init__(itself: float = 1, exchangeable: float = 0.8, mandatory: float = 1, accessory: float = 0.5, neutral: float = 0, out_of_cluster: float = 0.7) None
- __repr__()
Return repr(self).
- __setattr__(name, value)
Implement setattr(self, name, value).
- __weakref__
list of weak references to the object (if defined)
get_best_hit_4_func
- macsypy.hit.get_best_hit_4_func(function, hits, key='score')[source]
select the best Loner among several ones encoding for same function
score
i_evalue
profile_coverage
- Parameters:
function (str) – the name of the function fulfill by the hits (all hits must have same function)
hits (sequence of
macsypy.hit.ModelHitobject) – the hits to filter.key (str) – The criterion used to select the best hit ‘score’, i_evalue’, ‘profile_coverage’
- Returns:
the best hit
- Return type:
macsypy.hit.ModelHitobject
sort_model_hits
- macsypy.hit.sort_model_hits(model_hits)[source]
Sort
macsypy.hit.ModelHitper function- Parameters:
model_hits – a sequence of
macsypy.hit.ModelHit- Returns:
dict {str function name: [model_hit, …] }
compute_best_MSHit
get_best_hits
- macsypy.hit.get_best_hits(hits, key='score')[source]
If several hits match the same protein, keep only the best match based either on
score
i_evalue
profile_coverage
- Parameters:
hits ([
macsypy.hit.CoreHitobject, …]) – the hits to filter, all hits must match the same protein.key (str) – The criterion used to select the best hit ‘score’, i_evalue’, ‘profile_coverage’
- Returns:
the list of the best hits
- Return type:
[
macsypy.hit.CoreHitobject, …]
